A parameter server (PS) is used to process a large number of offline and online training tasks. SMART is short for scalable multiple additive regression tree. PS-SMART is an iteration algorithm that is implemented by using a PS-based gradient boosting decision tree (GBDT). The PS-SMART Binary Classification Training component supports training tasks for tens of billions of samples and hundreds of thousands of features. It can run training tasks on thousands of nodes. This component also supports multiple data formats and optimization technologies such as histogram-based approximation.
Limits
The input data of the PS-SMART Regression component must meet the following requirements:
Only columns of numeric data types can be used by the PS-SMART Regression component. If the type of data in the MaxCompute table is STRING, the data type must be converted first.
If data is in the key-value format, feature IDs must be positive integers, and feature values must be real numbers. If the data type of feature IDs is STRING, you must use the serialization component to serialize the data. If feature values are categorical strings, you must perform feature engineering such as feature discretization to process the values.
The PS-SMART Regression component can support hundreds of thousands of feature tasks. However, these tasks are resource-intensive and time-consuming. To resolve this issue, you can use GBDT algorithms to train the model. GBDT algorithms are suitable for the scenario where continuous features are used for training. You can perform one-hot encoding on categorical features to filter low-frequency features. However, we recommend that you do not perform feature discretization on continuous features of numeric data types.
The PS-SMART algorithm may introduce randomness. For example, randomness may be introduced in the following scenarios: data and feature sampling based on data_sample_ratio and fea_sample_ratio, optimization of the PS-SMART algorithm by using histograms for approximation, and merge of a local sketch into a global sketch. The structures of trees are different when jobs run on multiple workers in distributed mode. However, the training effect of the model is theoretically the same. It is normal if you use the same data and parameters during training but obtain different results.
If you want to accelerate training, you can set the Cores parameter to a larger value. The PS-SMART algorithm starts training tasks after the required resources are provided. Therefore, the more the resources are requested, the longer you must wait.
Precautions
When you use the PS-SMART Regression component, you must take note of the following items:
The PS-SMART Regression component can support hundreds of thousands of feature tasks. However, these tasks are resource-intensive and time-consuming. To resolve this issue, you can use GBDT algorithms to train the model. GBDT algorithms are suitable for the scenario where continuous features are used for training. You can perform one-hot encoding on categorical features to filter low-frequency features. However, we recommend that you do not perform feature discretization on continuous features of numeric data types.
The PS-SMART algorithm may introduce randomness. For example, randomness may be introduced in the following scenarios: data and feature sampling based on data_sample_ratio and fea_sample_ratio, optimization of the PS-SMART algorithm by using histograms for approximation, and merge of a local sketch into a global sketch. The structures of trees are different when jobs run on multiple workers in distributed mode. However, the training effect of the model is theoretically the same. It is normal if you use the same data and parameters during training but obtain different results.
If you want to accelerate training, you can set the Cores parameter to a larger value. The PS-SMART algorithm starts training tasks after the required resources are provided. Therefore, the more the resources are requested, the longer you must wait.
Configure the component
You can configure the PS-SMART Regression component by using one of the following methods:
Method 1: Configure the component in the Platform for AI (PAI) console
Configure the component parameters in Machine Learning Designer. The following table describes the parameters.
Tab | Parameter | Description |
Fields Setting | Use Sparse Format | Specifies whether the input data is in the sparse format. If data in the sparse format is presented by using key-value pairs, separate the key-value pairs with spaces and separate keys and values with colons (:). Example: 1:0.3 3:0.9. |
Feature Columns | Select the feature columns for training from the input table. If data in the input table is in the dense format, only the columns of the BIGINT and DOUBLE types are supported. If data in the input table is in the sparse format presented by using key-value pairs, and keys and values are of numeric data types, only columns of the STRING type are supported. | |
Label Column | Select the label column from the input table. The columns of the STRING type and numeric data types are supported. However, only data of numeric data types can be stored in the columns. For example, column values can be 0 or 1 in binary classification. | |
Weight Column | Select the column that contains the weight of each row of samples. The columns of numeric data types are supported. | |
Parameters Setting | Objective Function Type | Valid values:
|
Tweedie Distribution Index | The index of the relationship between the variance and mean value of Tweedie distribution. This parameter is supported only when you set the Objective Function Type parameter to Tweedie Regression. | |
Evaluation Indicator Type | Valid values:
| |
Trees | Enter the number of trees. The value of this parameter must be an integer. The number of trees is proportional to the amount of training time. | |
Maximum Tree Depth | The default value is 5, which specifies that up to 32 leaf nodes can be configured. | |
Data Sampling Fraction | Enter the data sampling ratio when trees are built. The sample data is used to build a weak learner to accelerate training. | |
Feature Sampling Fraction | Enter the feature sampling ratio when trees are built. The sample features are used to build a weak learner to accelerate training. | |
L1 Penalty Coefficient | Control the size of a leaf node. A larger value results in a more even distribution of leaf nodes. If overfitting occurs, increase the parameter value. | |
L2 Penalty Coefficient | Control the size of a leaf node. A larger value results in a more even distribution of leaf nodes. If overfitting occurs, increase the parameter value. | |
Learning Rate | Enter the learning rate. Valid values: (0,1). | |
Sketch-based Approximate Precision | Enter the threshold for selecting quantiles when you build a sketch. A smaller value specifies that more bins can be obtained. In most cases, the default value 0.03 is used. | |
Minimum Split Loss Change | Enter the minimum loss change required for splitting a node. A larger value specifies that node splitting is less likely to occur. | |
Features | Enter the number of features or the maximum feature ID. If this parameter is not specified for resource usage estimation, the system automatically runs an SQL task to calculate the number of features or the maximum feature ID. | |
Global Offset | Enter the initial prediction values of all samples. | |
Random Seed | Enter the random seed. The value of this parameter must be an integer. | |
Feature Importance Type | Valid values:
| |
Tuning | Cores | Select the number of cores. By default, the system determines the value. |
Memory Size per Core | Select the memory size of each core. Unit: MB. In most cases, the system determines the memory size. |
Method 2: Configure the component by using PAI commands
Configure the component parameters by using PAI commands. The following section describes the parameters. You can use the SQL Script component to call PAI commands. For more information, see SQL Script.
# Training
PAI -name ps_smart
-project algo_public
-DinputTableName="smart_regression_input"
-DmodelName="xlab_m_pai_ps_smart_bi_545859_v0"
-DoutputTableName="pai_temp_24515_545859_2"
-DoutputImportanceTableName="pai_temp_24515_545859_3"
-DlabelColName="label"
-DfeatureColNames="features"
-DenableSparse="true"
-Dobjective="reg:linear"
-Dmetric="rmse"
-DfeatureImportanceType="gain"
-DtreeCount="5"
-DmaxDepth="5"
-Dshrinkage="0.3"
-Dl2="1.0"
-Dl1="0"
-Dlifecycle="3"
-DsketchEps="0.03"
-DsampleRatio="1.0"
-DfeatureRatio="1.0"
-DbaseScore="0.5"
-DminSplitLoss="0"
# Prediction
PAI -name prediction
-project algo_public
-DinputTableName="smart_regression_input";
-DmodelName="xlab_m_pai_ps_smart_bi_545859_v0"
-DoutputTableName="pai_temp_24515_545860_1"
-DfeatureColNames="features"
-DappendColNames="label,features"
-DenableSparse="true"
-Dlifecycle="28"Module | Parameter | Required | Description | Default value |
Data parameters | featureColNames | Yes | The feature columns that are selected from the input table for training. If data in the input table is in the dense format, only the columns of the BIGINT and DOUBLE data types are supported. If data in the input table is in the sparse format presented by using key-value pairs, and keys and values are of numeric data types, only columns of the STRING data type are supported. | No default value |
labelColName | Yes | The label column in the input table. The columns of the STRING type and numeric data types are supported. However, only data of numeric data types can be stored in the columns. For example, column values can be 0 or 1 in binary classification. | No default value | |
weightCol | No | The column that contains the weight of each row of samples. The columns of numeric data types are supported. | No default value | |
enableSparse | No | Specifies whether the input data is in the sparse format. Valid values: true and false. Specifies whether the input data is in the sparse format. If data in the sparse format is presented by using key-value pairs, separate the key-value pairs with spaces and separate keys and values with colons (:). Example: 1:0.3 3:0.9. | false | |
inputTableName | Yes | The name of the input table. | No default value | |
modelName | Yes | The name of the output model. | No default value | |
outputImportanceTableName | No | The name of the table that provides feature importance. | No default value | |
inputTablePartitions | No | The partitions that are selected from the input table for training. Format: ds=1/pt=1. | No default value | |
outputTableName | No | The name of the table that is exported to MaxCompute. The table name must be in the binary format. | No default value | |
lifecycle | No | The lifecycle of the output table. Unit: days. | 3 | |
Algorithm parameters | objective | Yes | The type of the objective function. Valid values:
| reg:linear |
metric | No | The type of the evaluation metric in the training set, which is included in stdout of the coordinator in a logview. Valid values:
| No default value | |
treeCount | No | The number of trees. The value is proportional to the training time. | 1 | |
maxDepth | No | The maximum depth of a tree. Valid values: 1 to 20. | 5 | |
sampleRatio | No | The data sampling ratio. Value range: (0,1]. If this parameter is set to 1.0, sampling is not performed. | 1.0 | |
featureRatio | No | The feature sampling ratio. Value range: (0,1]. If this parameter is set to 1.0, no data is sampled. | 1.0 | |
l1 | No | The L1 penalty coefficient. A larger value of this parameter indicates a more even distribution of leaf nodes. If overfitting occurs, increase the parameter value. | 0 | |
l2 | No | The L2 penalty coefficient. A larger value of this parameter indicates a more even distribution of leaf nodes. If overfitting occurs, increase the parameter value. | 1.0 | |
shrinkage | No | The learning rate. Value range: (0,1). | 0.3 | |
sketchEps | No | The threshold for selecting quantiles when you build a sketch. The number of bins is O(1.0/sketchEps). A smaller value specifies that more bins can be obtained. In most cases, the default value is used. Value range: (0,1). | 0.03 | |
minSplitLoss | No | Enter the minimum loss change required for splitting a node. A larger value specifies that node splitting is less likely to occur. | 0 | |
featureNum | No | The number of features or the maximum feature ID. If this parameter is not specified for resource usage estimation, the system automatically runs an SQL task to calculate the number of features or the maximum feature ID. | No default value | |
baseScore | No | The initial prediction values of all samples. | 0.5 | |
randSeed | No | The random seed. The value of this parameter must be an integer. | No default value | |
featureImportanceType | No | The feature importance type. Valid values:
| gain | |
tweedieVarPower | No | The index of the relationship between the variance and mean value of Tweedie distribution. | 1.5 | |
Tuning parameters | coreNum | No | The number of cores that are used by the computing algorithm. The larger the value, the faster the computing algorithm runs. | Specified by the system |
memSizePerCore | No | The memory size of each core. Unit: MB. | Specified by the system |
Example
Execute the following SQL statements to generate input data on an ODPS SQL node. In this example, the input data in the key-value pairs is generated.
drop table if exists smart_regression_input; create table smart_regression_input as select * from ( select 2.0 as label, '1:0.55 2:-0.15 3:0.82 4:-0.99 5:0.17' as features union all select 1.0 as label, '1:-1.26 2:1.36 3:-0.13 4:-2.82 5:-0.41' as features union all select 1.0 as label, '1:-0.77 2:0.91 3:-0.23 4:-4.46 5:0.91' as features union all select 2.0 as label, '1:0.86 2:-0.22 3:-0.46 4:0.08 5:-0.60' as features union all select 1.0 as label, '1:-0.76 2:0.89 3:1.02 4:-0.78 5:-0.86' as features union all select 1.0 as label, '1:2.22 2:-0.46 3:0.49 4:0.31 5:-1.84' as features union all select 0.0 as label, '1:-1.21 2:0.09 3:0.23 4:2.04 5:0.30' as features union all select 1.0 as label, '1:2.17 2:-0.45 3:-1.22 4:-0.48 5:-1.41' as features union all select 0.0 as label, '1:-0.40 2:0.63 3:0.56 4:0.74 5:-1.44' as features union all select 1.0 as label, '1:0.17 2:0.49 3:-1.50 4:-2.20 5:-0.35' as features ) tmp;The following figure shows the generated training data.
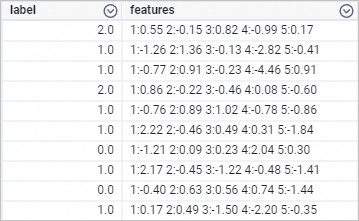
Create a pipeline shown in the following figure and run the component. For more information, see Algorithm modeling.
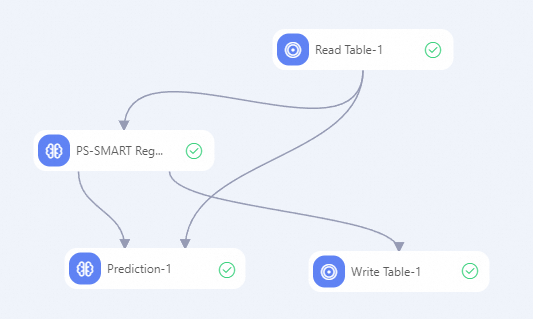
In the left-side component list of Machine Learning Designer, separately search for the Read Table, PS-SMART Regression, Prediction, and Write Table components, and drag the components to the canvas on the right.
Configure the component parameters.
On the canvas, click the Read Table-1 component. On the Select Table tab in the right pane, set Table Name to smart_regression_input.
On the canvas, click the PS-SMART Regression-1 component and configure the parameters in the right pane. The following table describes the parameters. Retain the default values for the parameters that are not described in the table.
Tab
Parameter
Description
Fields Setting
Use Sparse Format
Select Use Sparse Format.
Feature Columns
Select the features column.
Label Column
Select the label column.
Parameters Setting
Objective Function Type
Set the parameter to Linear Regression.
Evaluation Indicator Type
Set the parameter to Rooted Mean Square Error.
Trees
Set the parameter to 5.
On the canvas, click the Prediction-1 component and configure the parameters. The following table describes the parameters. Retain the default values for the parameters that are not described in the table.
Tab
Parameter
Description
Fields Setting
Feature Columns
By default, all columns in the input table are selected. Specific columns may not be used for training. These columns do not affect the prediction result.
Reserved Columns
Select the label column.
Sparse Matrix
Select Sparse Matrix.
KV Delimiter
Set this parameter to a colon (:).
KV Pair Delimiter
Set this parameter to a space.
On the canvas, click the Write Table-1 component. On the Select Table tab in the right pane, set New Table Name to smart_regression_output.
After the parameter configuration is complete, click the
 button to run the pipeline.
button to run the pipeline.
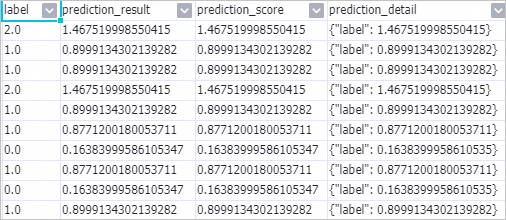
Right-click the Prediction-1 component and choose to view the prediction results.
Right-click the PS-SMART Regression-1 component and choose to view the feature importance table.
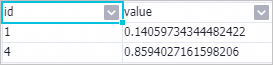
id: the ID of a passed feature. In this example, the input data is in the key-value format. The values in the id column indicate the keys in the key-value pairs. The feature importance table has only two features. This indicates that only these two features are used in the split of the tree. The feature importance of other features can be considered as 0. value: the feature importance type. The default value of this parameter is gain, which indicates the sum of the information gains provided by the feature for the model.
PS-SMART model deployment
If you want to deploy the model generated by the PS-SMART Binary Classification Training component to EAS as an online service, you must add the Model export component as a downstream node for the PS-SMART Binary Classification Training component and configure the Model export component. For more information, see Model export.
After the Model export component is successfully run, you can deploy the generated model to EAS as an online service on the EAS-Online Model Services page. For more information, see Deploy a model service in the PAI console.
References
For more information about Machine Learning Designer, see Overview of Machine Learning Designer.
Machine Learning Designer provides various preset algorithm components. You can use a component to process data based on your business requirements. For more information, see Component reference: Overview of all components.