Bolster the growth and digital transformation of your business amid the outbreak through the Anti COVID-19 SME Enablement Program. Get a $300 coupon package for all new SME customers or a $500 coupon for paying customers.
By ApsaraDB.
Towards the end of last year, a mysterious disease emerged in Wuhan, an emerging commercial center in central China. In a matter of a few months, the disease resulted in well over 3,000 deaths and more than 82,000 confirmed case in China. Since then, the disease has become all too familiar to the world, as the world is facing a pandemic. Affecting nearly 200 countries, with hundreds of thousands of deaths and around two million confirmed cases, and trillions of dollars in economic losses worldwide. In the face of this grave situation, Alibaba Cloud has made it our duty to help prevent and control the pandemic through technological innovations. As a part of this, our AnalyticDB team has provided an efficient gene sequence retrieval system to assist in COVID-19 virus sequence analysis.
As of now, the technology involved in gene sequencing and analysis is mainly used for the following uses. The first use is to trace and analyze the SARS-CoV-2 virus to help in contact tracing and other effective prevention and control measures. Through gene matching technology, we found that the SARS-CoV-2 RNA sequence has a 96% match with other coronaviruses that typically infect bats and a 99.7% match to coronaviruses that infect pangolins. This means that pangolins and bats were likely culprits for the original hosts of the coronavirus. The second use of gene sequence analysis is to divide the functional parts of the gene sequence to understand the function of each module; doing this allows us to analyze how the virus is replicated and transmitted. This is important because, once we identify the key nodes, we can design targeted drugs and vaccines. The third use is to aid drug treatment and therapy research. In particular, researchers can retrieve the gene sequences of diseases similar to COVID-19, such as SARS and MERS, and look into treatments for these similar diseases. Doing so means that test kits, vaccines and therapeutic drugs can be designed and developed more quickly and efficiently.
However, the current gene matching techniques used in the industry are inefficient. Therefore, it is urgent that we have an efficient matching algorithm for gene sequence analysis. Alibaba Cloud's AnalyticDB team has been converting gene fragments into 1024-dimensional vector features, which allows us to convert the problem of matching two gene fragments into a calculation of the distance between vectors, greatly reducing the computing overhead and allowing the computer to return the result in milliseconds. This process can be used for preliminary gene fragment screening. For gene similarity calculation, we use the Blast algorithm[5] to output a precise similarity ranking. This completes the gene sequence matching calculation. Alibaba Cloud AnalyticDB also provides powerful machine learning analysis tools. These tools can convert local and disease-related target gene fragments into feature vectors by using gene-to-vector technology. These vectors can then be used in the design of gene drugs, greatly accelerating the process of genetic analysis.
The RNA sequence of COVID-19 can be expressed as a string of nucleic acid sequences, which is also referred to as base sequences. The RNA sequence is made up of four nucleotides, labeled A, C, G, and T for adenine, cytosine, guanine, and thymine. Each letter represents a base, and these bases are linked together without gaps. Each virus has a different RNA sequence. The gene retrieval system allows users to input a virus gene fragment to search for similar genes, which is used to process the virus RNA.
To demonstrate our approach to gene fragment retrieval, we downloaded a large number of viral RNA fragments from GenBank and then also imported virus-related papers from GenBank and Google Scholar into the AnalyticDB gene retrieval database.
The gene retrieval demo UI is shown in Figure 1 below. First, the user uploads the COVID-19 sequence to AnalyticDB's gene search tool. Then, the system retrieves similar gene fragments in a few milliseconds. Currently, the system only returns gene fragments with a matching degree greater than 0.8. For this, the Guangdong pangolin coronavirus (GD/P1L), bat coronavirus (RaTG13), SARS, and MARS are returned. As you can see, GD/P1L is the best sequence match with a matching degree of 0.9979. This tells us that COVID-19 was likely transmitted to people through pangolins.
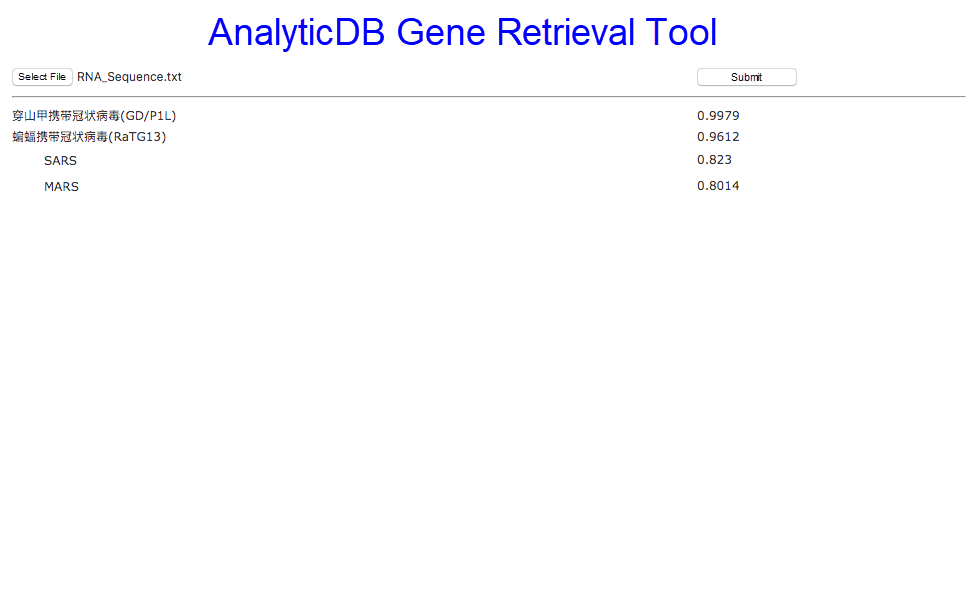
Figure 1. Gene retrieval interface
In a case where RNA fragments are very similar, it means that the two RNA sequences may have similar protein expressions and structures. By using the gene retrieval tool, we can see that the matching degree between SARS and MARS and COVID-19 is greater than 0.8. This indicates that we may be able to use some of previous research studies concerning SARS or MARS to better understand COVID-19. After obtaining matching viruses, the system crawls the academic papers about each of the matching viruses and divides these papers into the testing, vaccine, and medication categories. If we click SARS shown in Figure 2, shown below, we can see that there are seven SARS testing methods, four vaccine production methods, and ten drug therapy methods. You can find that one of the testing methods for SARS is fluorescence quantitative PCR detection, which is being used in COVID-19 testing. Looking at the vaccine literature, we can note that the gene vaccine and in vivo induction of immune response methods proposed for SARS are also being explored for COVID-19. From the drug literature, Remdesivir and other relevant interferons, a category of antivirals, are currently being used to treat COVID-19 patients.
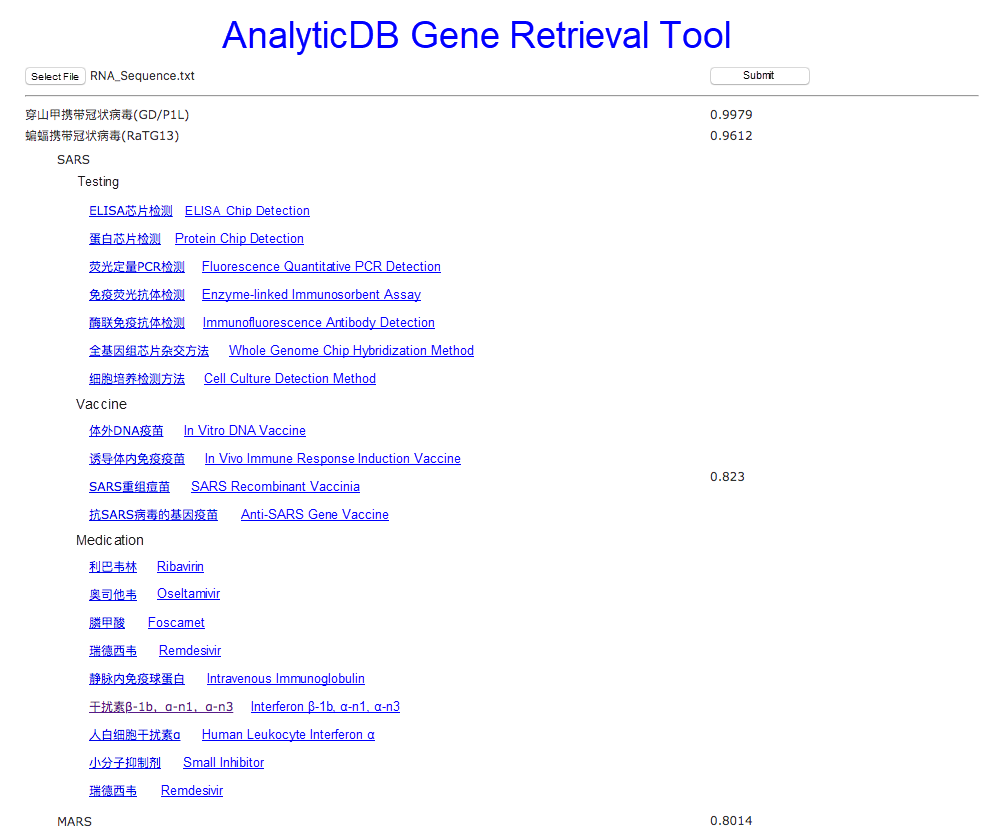
Figure 2. Literature classification
If we click the link to the interferon paper, the relevant paper appears, as shown in Figure 3. At present, the system uses automatic translation software to translate the papers and extracts keywords from the Chinese file names to translate the file names. This makes it easier for non-Chinese readers to understand the materials.
Figure 3. Click the interferon paper link
The overall architecture of the Alibaba Cloud gene retrieval system is shown in Figure 4. In this architecture, AnalyticDB is responsible for processing all of the application's structured data, such as gene sequence lengths, academic paper names that contain the gene, gene types, DNA/RNA. This is shown in the query result part of Figure 4. AnalyticDB is also responsible for the storage and query of the feature vectors produced for gene sequences. During the query process, we use a gene vector extraction model to convert genes into vectors and perform coarse sorting retrieval in AnalyticDB. In the vector matching result set, we use the classic BLAST[7] algorithm to perform precise sorting and return the most similar gene sequences.
Most importantly, the gene vector extraction module contains the vectors derived from nucleotide sequences. So far, we have extracted all the sequence samples of various viral RNA for training to help the model better calculate the similarity of viral RNA. Undoubtedly, the current vector extraction model can be easily extended to genes of other species. The gene vector extraction model is described in detail in section 3.
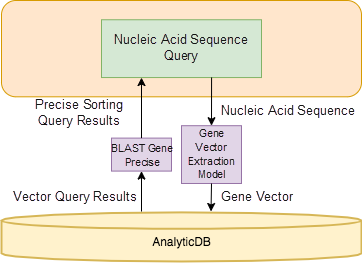
Figure 4. Gene retrieval framework
To better explain gene vector extraction, let's look at a word vector extraction algorithm, which follows the same principles. Word vector1 technology is already highly sophisticated. It is widely used in machine translation, reading comprehension, semantic analysis, and other related fields, where it has achieved great success. Word vectorization uses a distributional semantic approach to express the meaning of a word. Here, the meaning of a word is its context. For example, think back to tests where you had to use the words in a wordbank to fill in missing words in a paragraph. In these tests, the context of a word can accurately reflect the word itself. By choosing the correct word, we show that we understand the meaning of the vacant word. Therefore, by using the relationship of a given word with surrounding words, a word vector algorithm can generate a vector for each word in a text. Then, we can calculate the similarity of word vectors to obtain the similarity between words. For example, the words "spoon" and "bowl" have a high similarity because they always appear in the context of eating.
Similarly, the arrangement of gene sequences follows certain rules, and each part of a gene sequence expresses different functions and meanings. Therefore, we can divide a long gene sequence into smaller units ("words") for research purposes. These "words" also have a context, because they are interconnected and interact with each other to complete corresponding functions and form reasonable expressions. Therefore, biological scientists7[9] used word vector algorithms to vectorize gene sequence units. A high similarity between two gene units indicates that both gene units always appear together and jointly express a corresponding function.
In short, vector extraction involves three main steps:
First, we must figure out how to define each "word" in an amino acid sequence. In the bioinformatics field, researchers use k-mers[3] to analyze amino acid sequences. K-mers are obtained by dividing a nucleic acid sequence into strings that contain K bases. This is done by iteratively selecting a sequence of K bases in length from a continuous nucleic acid sequence. If the length of the nucleic acid sequence is L, we can obtain L-K+1 k-mers from it. As shown in Figure 5, assuming that the length of a sequence is 12, if we set the k-mer length to 8, five (12-8+1=5) 8-mers can be obtained. These k-mers are exactly the "words" in the amino acid sequence.
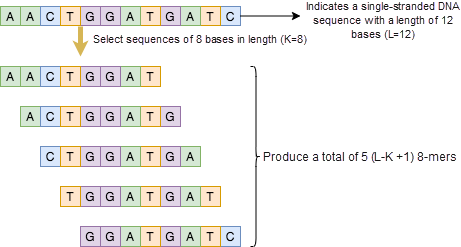
Figure 5. Diagram of the 8-mer nucleic acid sequence
Second, the context plays an important role in word vector algorithms. For amino acid fragments, we choose a window with a length of L, and the amino acid fragments in this window are considered to be within the same context. For example, if we select a window with a length of 10 for the nucleic acid sequence CTGGATGA, we can convert it into five 5-mers: AACTG, ACTGG, CTGGA, GGATG, and GATGA. When we select one 5-mer (CTGGA), then the other four 5-mers (AACTG, ACTGG, GGATG, ad GATGA) compose its context. By using a word vector space training model and performing training on the existing genetic k-mers of different organisms, we can convert a k-mer (a "word" in a gene sequence) into a 1024-dimensional vector.
Third, similar to word vector models; k-mer vector models also perform mathematical computations on vectors.
Vector subtraction: 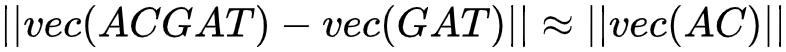 (1)
(1)
Vector addition: 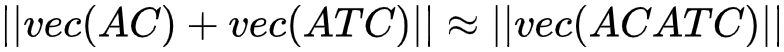 (2)
(2)
Formula 1 indicates that the distance between "ACGAT vector minus AC vector" and the GAT vector is very close. Formula 2 shows that the distance between "AC vector plus ATC vector" and the ACATC vector is also very close. Based on these mathematical characteristics, when we want to calculate the vector of a long amino acid sequence, we can add the k-mer sequences of each fragment into this sequence. Then, by normalizing the result, we can obtain the vector of the whole amino acid sequence. To further improve the accuracy of this approach, we can take a gene fragment as a text fragment and use doc2vec4 to convert the entire sequence into a vector for calculation.
To verify the performance of the algorithm, we calculated the similarity between the BLAST[6] algorithm sequence and the vector-to-gene l2 distance sequence. The Spearman rank correlation coefficient[6] of both sequences was 0.839. This shows that converting DNA sequences into vectors is an effective means of preliminary screening for similar gene fragments.
AnalyticDB is a high-concurrency, low-latency, and real-time data warehouse on Alibaba Cloud, which supports petabytes of data. It supports instant multi-dimensional analysis and service exploration for trillions of data entries within milliseconds.
AnalyticDB for MySQL is fully compatible with the MySQL protocol and the SQL:2003 standard. AnalyticDB for PostgreSQL supports SQL:2003 and is highly compatible with the Oracle syntax ecosystem. Currently, both products provide a vector search function, which supports similarity queries for images, recommendations, voiceprints, and nucleotide sequences. In actual application scenarios, AnalyticDB can query billions of vector data entries and respond within 100 milliseconds. AnalyticDB has been widely used in security projects across multiple cities.
In general application systems that involve vector search, developers usually use a vector search engine, such as Faiss, to store vector data and then use relational databases to store structured data. This means you have to alternate between both systems during queries. Moreover, this solution requires extra development efforts and does not provide optimal performance.
AnalyticDB supports the retrieval of structured and non-structured data (vectors). This means you can simply use an SQL interface to quickly implement gene retrieval and hybrid gene + structured data retrieval. In hybrid retrieval scenarios, the optimizer of AnalyticDB selects the optimal execution plan based on the data distribution and query conditions in order to achieve optimal performance while ensuring retrievability.
RNA nucleic acid sequence retrieval can be implemented with one SQL statement:
-- 查找RNA和提交的序列向量相近的基因序列。
select title, # 文章名
length, # 基因长度
type, # mRNA或DNA等
l2_distance(feature, array[-0.017,-0.032,...]::real[]) as distance # 向量距离
from demo.paper a, demo.dna_feature b
where a.id = b.id
order by distance; # 用向量相似度排序The demo.paper table stores the basic information of each uploaded article, and demo.dna_feature stores the vectors that correspond to gene sequences of each species. The gene-to-vector model is used to convert genes to vectors like [-0.017,-0.032,...] so that they can be used for retrieval in Alibaba Cloud AnalyticDB.
The current system also supports structured information and non-structured information (nucleotide sequence) hybrid retrieval. For example, assume we want to search for gene segments that are similar to the SARS-CoV-2 virus. To use AnalyticDB, you only need to add the substatement "where title like'%COVID-19%'" in the SQL statement to easily perform this search.
[1] Mikolov Tomas; et al. (2013). "Efficient Estimation of Word Representations in Vector Space". arXiv:1301.3781
[2] Mikolov Tomas, Sutskever Ilya, Chen Kai, Corrado, Greg S., and Dean Jeff (2013). Distributed representations of words and phrases and their compositionality. Advances in Neural Information Processing Systems. arXiv:1310.4546. Bibcode:2013arXiv1310.4546M.
[3] Mapleson Daniel, Garcia Accinelli, Gonzalo, Kettleborough George, Wright Jonathan and Clavijo, Bernardo J. (2016). "KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies". Bioinformatics. 33(4): 574–576. doi:10.1093/bioinformatics/btw663. ISSN 1367-4803. PMC 5408915. PMID 27797770.
[4] Quoc Le and Tomas Mikolov. (2014). Distributed representations of sentences and documents. In International Conference on Machine Learning, pages 1188–1196.
[5] Stephen F Altschul, Warren Gish, Webb Miller, Eugene W. Myers, and David J. Lipman: Basic local alignment search tool. (1990), Journal of Molecular Biology, 215(3):403-410.
[6] Julia Piantadosi, Phil Howlett, and John Boland. (2007). "Matching the grade correlation coefficient using a copula with maximum disorder", Journal of Industrial and Management Optimization, 3 (2), 305–312
[7] Stephen Woloszynek, Zhengqiao Zhao, Jian Chen, and Gail L. Rosen. (2019). "16s rRNA sequence embeddings: Meaningful numeric feature representations of nucleotide sequences that are convenient for downstream analyses", PLoS Computational Biology, 15(2), e1006721.
[8] James K. Senter, Taylor M. Royalty, Andrew D. Steen, and Amir Sadovnik. (2019) "Unaligned Sequence Similarity Search Using Deep Learning.", arXiv e-prints
[9] Ng Patrick. (2017) dna2vec: consistent vector representations of variable-length k-mers. arXiv preprint, arXiv: 1701.06279.
While continuing to wage war against the worldwide outbreak, Alibaba Cloud will play its part and will do all it can to help others in their battles with the coronavirus. Learn how we can support your business continuity at https://www.alibabacloud.com/campaign/fight-coronavirus-covid-19

2,593 posts | 793 followers
FollowAlibaba Clouder - June 5, 2020
Alibaba Clouder - June 12, 2020
Alibaba Clouder - April 30, 2020
Alibaba Clouder - April 24, 2020
Alibaba Clouder - April 29, 2020
Alibaba Clouder - March 6, 2020

2,593 posts | 793 followers
Follow Platform For AI
Platform For AI
A platform that provides enterprise-level data modeling services based on machine learning algorithms to quickly meet your needs for data-driven operations.
Learn More MaxCompute
MaxCompute
Conduct large-scale data warehousing with MaxCompute
Learn More AnalyticDB for MySQL
AnalyticDB for MySQL
AnalyticDB for MySQL is a real-time data warehousing service that can process petabytes of data with high concurrency and low latency.
Learn MoreMore Posts by Alibaba Clouder